
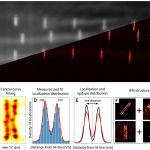
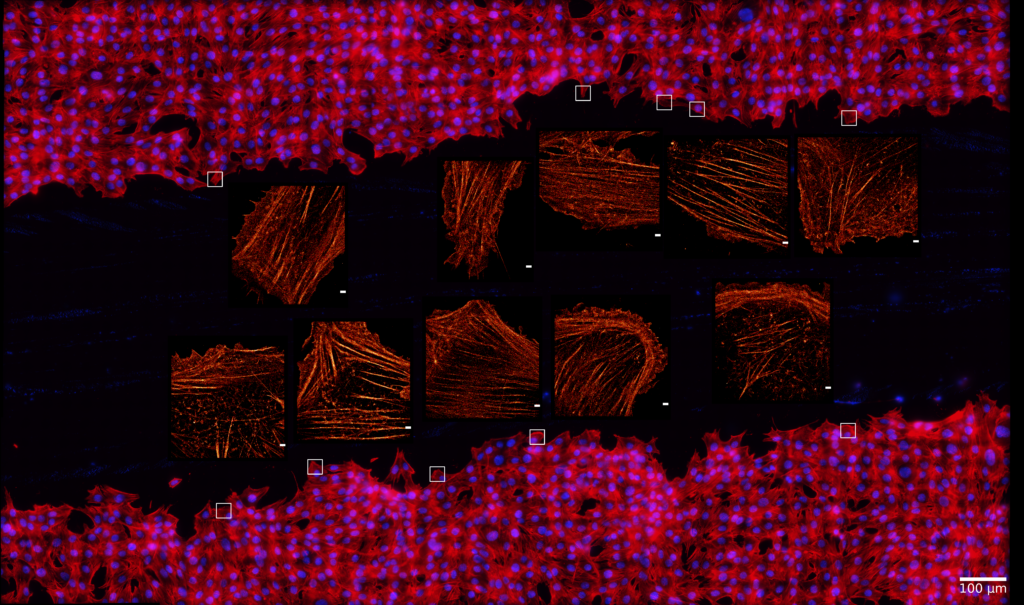
In standard SMLM methods, the photoswitching of single fluorescent molecules and the data acquisition processes are independent, which leads to the detection of single molecule blinking events on several consecutive frames. This mismatch results in several data points with reduced localization precision, and it also increases the possibilities of overlapping. Here we discuss how the synchronization of the fluorophores’ ON state to the camera exposure time increases the average intensity of the captured point spread functions and hence improves the localization precision. Simulations and theoretical results show that such synchronization leads to fewer localizations with 15% higher sum signal on average, while reducing the probability of overlaps by 10%.












You must be logged in to post a comment.